Research in the Schroeder lab is driven by a fascination with the vast, largely unexplored universe of biogenic small molecules (BSMs). Recent advances in mass spectrometry (MS) have revealed >10,000 compounds in the metabolomes of animal model systems; however, the chemical structures of most detected compounds have not been elucidated and their biological functions remain unknown.
Our work has shown that previously unidentified BSMs control all aspects of biology, including neurotransmitter signaling, brain development in mouse, fat storage, as well as many aspects of development and longevity. We are particularly interested in the role of microbiota - all animals are associated with cemomplex microbiomes that produce BSMs with profound effects on the host.
We use two model systems: the nematode model C. elegans and mouse. C. elegans is an extremely "chemist-friendly" model system; it can be grown easily in the lab, like bacteria, for biochemical studies, its genome is extremely well characterized (better than that of human or mouse), the genome can be edited "at will" to introduce all sorts of non-natural features to enable chmeical biology, and nonetheless features a highly differentiated physiology, and it has become apparent that many signaling pathways in C. elegans show strong analogies to corresponding pathways in higher animals.
Our C. elegans work provided the foundation for our investigations of the role of microbiota for mouse and human biology. In collaboration with Davbid Artis at Weill Cornell we have shown that microbiota-dependent metabolites are required for normal brain development and immuneregulation. Ongoing work indicates that fat storage and high cholesterol are in large measure regulated by metabolites of "mixed" orgin, i.e. BSMs whose biosynthesis involves both the host and the microbiota.
Research Highlights
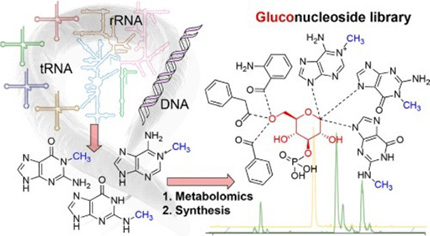
Gluconucleosides from RNA
Nucleosides are essential cornerstones of life, and nucleoside derivatives and synthetic analogues have important biomedical applications. We recently reported the discovery of glucose-based nucleosides in Caenorhabditis elegans. Using a mass spectrometric screen based on all-ion fragmentation in combination with total synthesis, we show that C. elegans selectively glucosylates modified purines, but not canonical purine and pyrimidine bases. Analogous to ribonucleosides, the resulting gluconucleosides exist as phosphorylated and non-phosphorylated forms. Phosphorylated gluconucleosides can be additionally decorated with diverse acyl moieties from amino acid catabolism. Syntheses of representative variants, facilitated by a novel phosphoryl transesterification reaction, demonstrated selective incorporation of different nucleobases and acyl moieties. Using stable-isotope labeling, we further show that gluconucleosides incorporate modified nucleobases derived from RNA and possibly DNA breakdown, revealing extensive recycling of oligonucleotide catabolites. Gluconucleosides are conserved in other nematodes, and we are currently investigating their biologucal functions. Check out our recent paper in JACS.
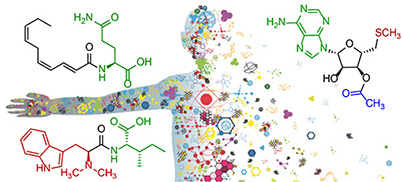
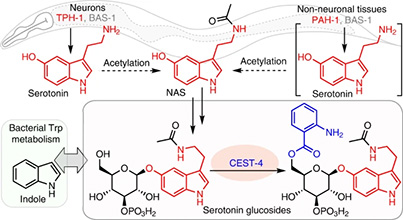
The origin and fate of serotonin
The neurotransmitter serotonin plays a central role in animal behavior and physiology, and many of its functions are regulated via evolutionarily conserved biosynthesis and degradation pathways. Here we show that in C. elegans, serotonin is abundantly produced in nonneuronal tissues via phenylalanine hydroxylase, in addition to canonical biosynthesis via tryptophan hydroxylase in neurons. Combining CRISPR–Cas9 genome editing, comparative metabolomics and synthesis, we demonstrate that most serotonin is further converted intoN-acetylserotonin-derived glucosides, which are retained in the animal and further modified via the carboxylesterase CEST-4. Expression patterns of CEST-4 suggest that serotonin or serotonin derivatives are transported between different tissues. Last, we show that bacterial indole production interacts with serotonin metabolism via CEST-4. Our results reveal a parallel pathway for serotonin biosynthesis in nonneuronal cell types and further indicate that serotonin-derived metabolites may serve distinct signaling functions and contribute to previously described serotonin-dependent phenotypes. Check out our recent paper in Nature Chemical Biology.
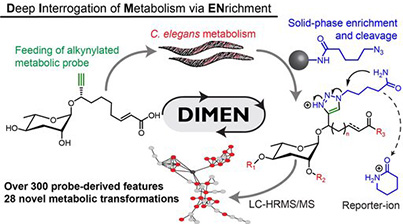
Deep Interrogation of Metabolism via Enrichment: DIMEN
Untargeted metabolomics indicates that the number of unidentified small-molecule metabolites may exceed the number of protein-coding genes for many organisms, including humans, by orders of magnitude. Uncovering the underlying metabolic networks is essential for elucidating the physiological and ecological significance of these biogenic small molecules. Here we develop a click-chemistry-based enrichment strategy, DIMEN (deep interrogation of metabolism via enrichment), that we apply to investigate metabolism of the ascarosides, a family of signaling molecules in the model organism C. elegans. Using a single alkyne-modified metabolite and a solid-phase azide resin that installs a diagnostic moiety for MS/MS-based identification, DIMEN uncovered several hundred novel compounds originating from diverse biosynthetic transformations that reveal unexpected intersection with amino acid, carbohydrate, and energy metabolism. Many of the newly discovered transformations could not be identified or detected by conventional LC-MS analyses without enrichment, demonstrating the utility of DIMEN for deeply probing biochemical networks that generate extensive yet uncharacterized structure space. Check out our DIMEN paper in JACS.
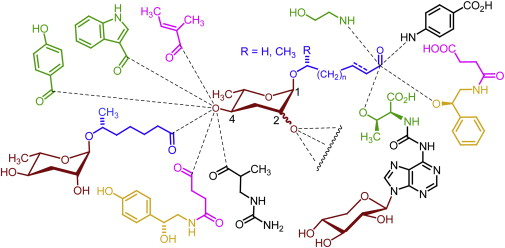
A combinatorial library of signaling molecules from the model organisms C. elegans and P. pacificus.
Toward a complete annotation of the C. elegans, mouse, and human metabolomes
Our efforts towards characterizing the C. elegans metabolome rely on innovative comparative metabolomics using 2D NMR and high-resolution MS/MS (see our recent paper in Nature Communications). This strategy greatly accelerates the process of identifying new chemical structures and elucidating their biological functions, and takes advantage of the extensive mutant and RNAi libraries that, in the case of C. elegans, cover almost the entire genome. Our human, mouse, and C. elegans metabolomics projects integrates analytical chemistry, synthetic chemistry, molecular biology, and protein biochemistry, and relies on an extensive network of collaborators at Caltech, Weill Cornell, MIT, and several Max Planck Institutes.
Pairing genome editing with metabolomics: new antibiotics from fungi
Fungi are an important part of the human microbiome but also among the most prolific sources of pharmacologically relevant natural products. Bioinformatic analyses of fungal genomes have revealed 1000s of biosynthetic gene clusters (BGCs), of which ~40% contain NRPS (non-ribosomal peptide synthetase) or NRPS-like genes. However, only a fraction of the biosynthetic capabilities of fungi have been discovered, because expression of many, perhaps even most biosynthetic pathways depends strongly on environmental conditions, including the presence of specific elicitors derived from the presence of other organisms. As a result, most fungal BGCs have no known metabolites (orphan BGCs). To address this challenge, our research leverages major recent advances in heterologous gene expression and comparative metabolomics, aiming to develop an innovative discovery pipeline for the systematic annotation of the biosynthetic capabilities of fungi. In collaboration with the lab of Nancy Keller at the University of Wisconsin-Madison, we are using a range of genome editing approaches to de-orphanize gene clusters and investigate newly uncovered biosynthetic pathways. For rapid chemical characterization of novel metabolites, we are combining concepts of comparative metabolomics, high-resolution MS/MS, and high dynamic range 2D NMR spectroscopy to develop a fast innovative structure elucidation process that is specifically tailored for surveying arrays of metabolite extracts generated from mutant libraries. See our recent paper in Nature Communications.
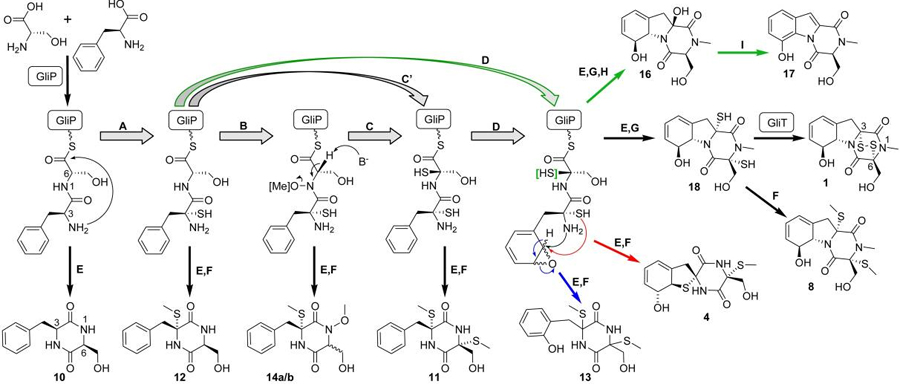
Putative biosynthetic pathway of the fungal metabolite gliotoxin, as inferred from 2D NMR-based comparative metabolomics.